Quantitative Imaging in Pathology (QuIP)
Description
QuIP is a web accessible toolset designed to support analysis, management, and exploration of whole slide tissue images for cancer research.
The QuIP system consists of a set of docker containers, which provide data management backend services, and web applications to load and visualize whole slide tissue images (in OpenSlide supported formats), run nuclear segmentation analyses on image tiles, and visualize and explore the analysis results.
The Application Service container hosts several web applications to view whole slide tissue images, view and interact with analysis results, and edit and curate analysis results. caMicroscope, for example, facilitates the interactive exploration of whole slide tissue images and analysis results (overlaid on images as polygons or heatmaps). FeatureScape enables exploration of imaging features computed for segmented nuclei. It supports visualizations such as scatter plots to allow one to take a deep dive into nucleus-level features.
| Viewing Classification Results | Viewing Segmentation Results |
|---|---|
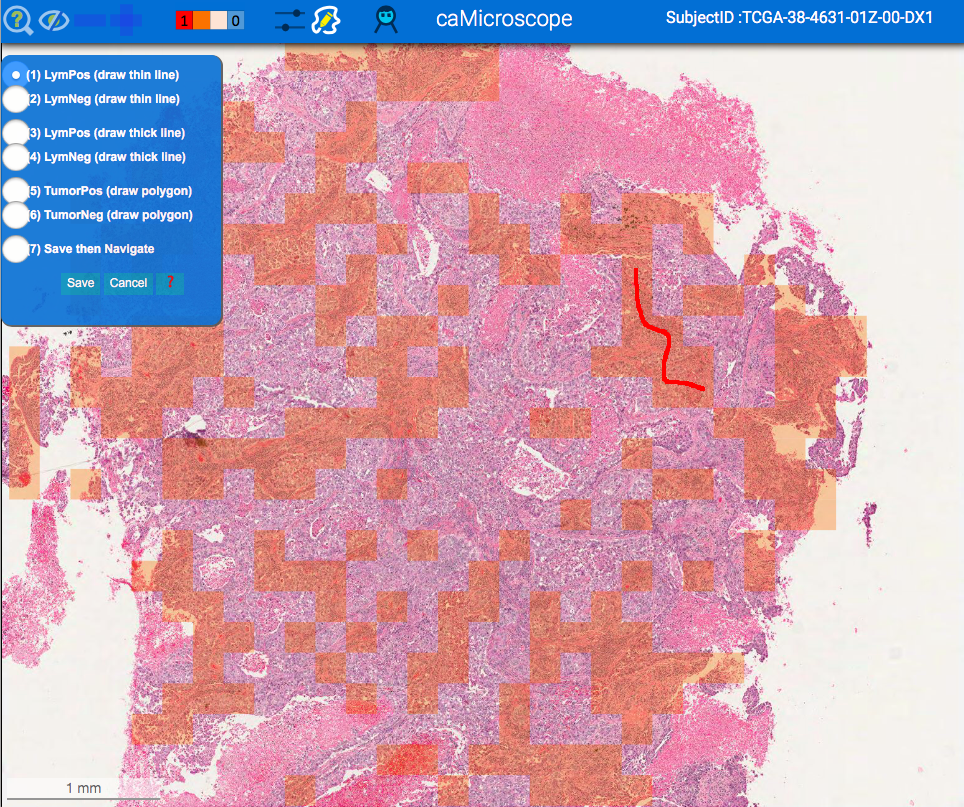 |
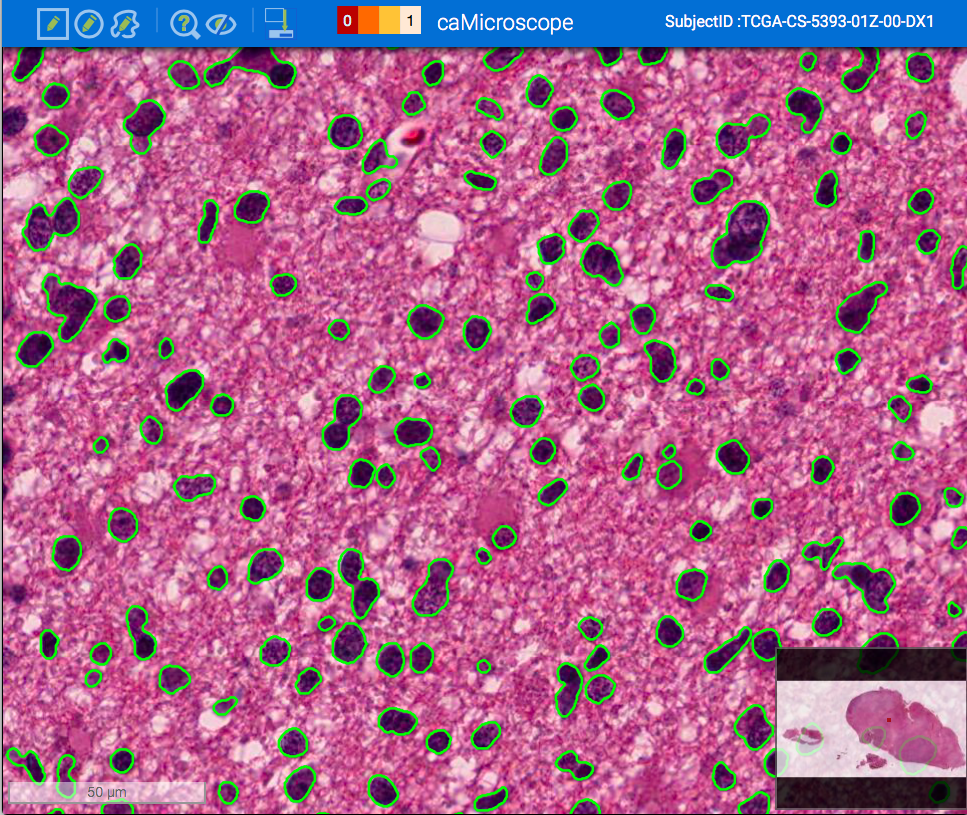 |
| Curation of Imaging Features | Feature Exploration via FeatureScape |
|---|---|
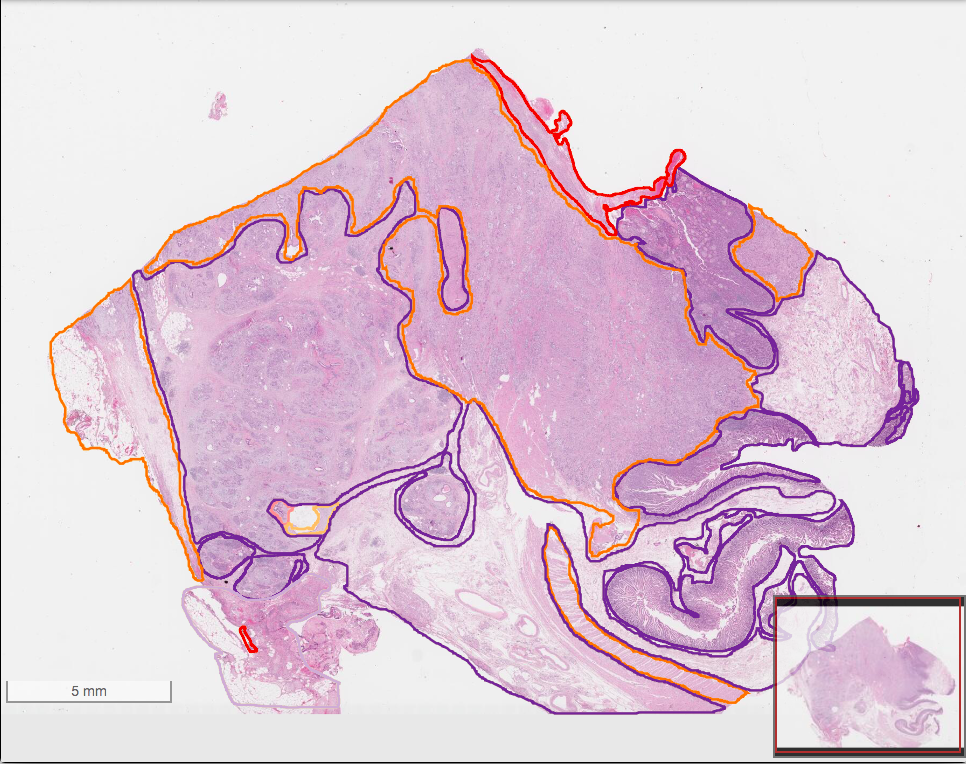 |
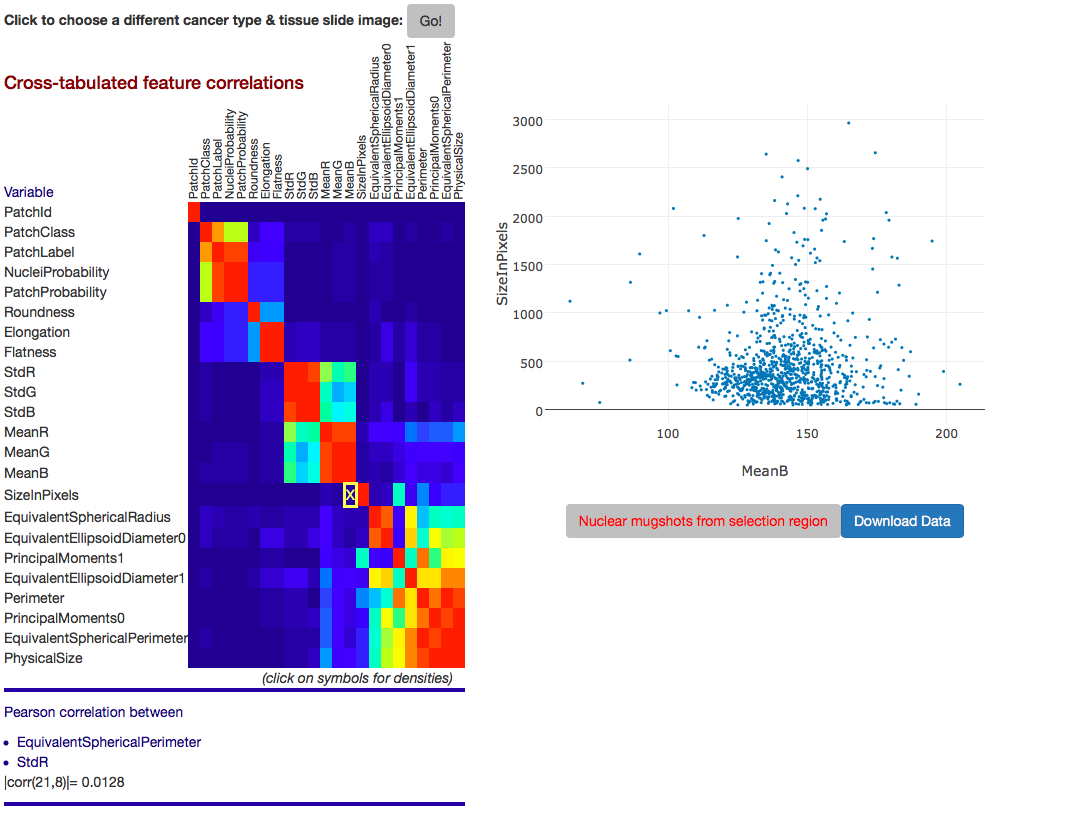 |
The Data Service containers implement (1) the database for storing image metadata, analysis results, and metadata about analyses (e.g., analysis parameters). The database organizes analysis results using a GeoJSON compliant specification. Segmentation results are expressed as polygons and size, shape, intensity and texture features, such as area, mean intensity, for each segmented object as key-value pairs; (2) the functions for loading image metadata and image analysis results, which are output from the analysis service as image masks and csv files which contain the computed features and boundaries of segmented objects; and (3) the functions for querying feature results by the FeatureScape web application.
Installing and Running QuIP
Requirements
- docker should be installed. You should be able to run docker commands (e.g., pull, run).
- git should be installed.
- QuIP requires access to a folder (data folder) on the host machine to store whole slide tissue images and the database. You should have read and write permissions to the folder.
- The installation scripts and the QuIP software have been tested on Linux systems and with the Google Chrome browser.
- Port 80 should be open to access and use the web applications remotely.
QuIP distribution repository
Clone this repository.
git clone https://github.com/SBU-BMI/quip_distro
Citation
When using QuIP or caMicroscope, please cite the following:
Saltz, J., Sharma, A., Iyer, G., Bremer, E., Wang, F., Jasniewski, A., DiPrima, T., Almeida, J.S., Gao, Y., Zhao, T., et al. (2017). A Containerized Software System for Generation, Management, and Exploration of Features from Whole Slide Tissue Images. Cancer Res. 77, e79–e82.
caMicroscope — A Platform for Digital Pathology Data Management and Visualization http://camicroscope.org